Microbial water quality standards are based upon indicator bacteria – fecal coliform or Escherichia coli. It is assumed that these bacteria can “indicate” the presence of pathogens, and associated human health risks. Pathogens of concern in rangeland streams include Cryptosporidium parvum, Salmonella, Campylobacter and Escherichia coli O157:H7. Microbial pollution sources include wildlife, humans, and livestock. In California, fecal coliform standards range from 20 to 2000 colony forming units (cfu) per 100 milliliter (mL) of water sampled. USEPA recommends an E. coli standard of 126 cfu/100ml for an average of 5 samples collected over 30 days, or 235 cfu/100mL for a single grab sample.
Objective
There is limited information to test the assumption that indicator bacteria correlate with the occurrence of pathogens in rangeland streams. We evaluated how well indicator bacteria predicted the occurrence of several waterborne pathogens at stream sites associated with major summer cattle grazing regions of the Sierra Nevada.
Study Sites
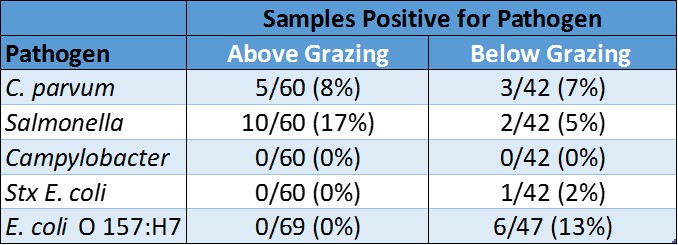
Nineteen sample locations were selected on streams above and below three major irrigated meadow systems. Meadow systems included Bridgeport Valley, Sierra Valley, and Goodrich Creek. Each meadow receives streamflow from surrounding mountains, streamflow is diverted to irrigate meadows for forage production, and meadows are grazed by beef cattle and in some cases cut for hay. Beef cattle numbers (animal units) for each site are approximately 20,000; 15,000; and 1,500 respectively. These sites are grazed from April through October.
We measured indicator bacteria and pathogens at stream sites above and below major summer livestock regions in the Sierra Nevada in order to evaluate correlations between microbial water quality standards and pathogens
Data Collection
From April through October of 2007 and 2008, we collected monthly samples at all 19 stream locations. In 2007, samples were transported to UC Davis for immediate analysis for fecal coliform and indicator E. coli (cfu/100mL), the protozoa pathogen C. parvum (oocysts/L), the bacterial pathogens Salmonella (MPN/100mL), Campylobacter (MPN/100mL) and Stx 1 and Stx2 E. coli (presence/absence). In 2008, samples were transported to UC Davis for immediate analysis for fecal coliform and indicator E. coli (cfu/100mL), and the bacterial pathogen E. coli O157:H7 (presence/absence).
Results
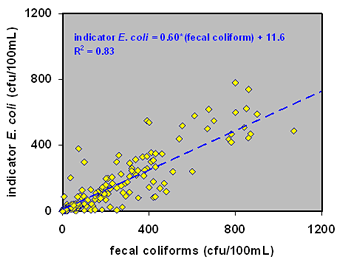
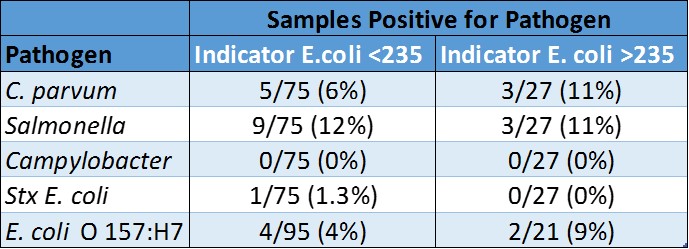
We found a strong correlation between indicator E. coli and fecal coliform, which is expected because indicator E. coli is a type of fecal coliform. However, the regulatory assumption that fecal coliforms and indicator E. coli are indicative of the presence of pathogens was not supported by our findings.
Management Implications
These results indicate that water quality regulation and monitoring based upon indicator bacteria may lead to unnecessary management restrictions. Alternatively, regulation based solely upon indicator bacteria may lead to a false sense of human health protection. Given the importance of this topic, consideration should be given to water quality monitoring and standards targeting specific microbial pathogens of concern.